ESR1 – Exploiting an innovative proteomic approach for novel pseudomonas aeruginosa vaccine antigen identification
Organisation: University College Dublin (Ireland) Researcher: Irene Jurado Supervisor: Assoc. Prof. Siobhán McClean Project description: P. aeruginosa (PA) is a major cause of opportunistic infections in hospitalised patients and in people with chronic pulmonary disorders (e.g. cystic fibrosis; CF), and chronic obstructive pulmonary disease (COPD). It is highly antibiotic resistant and there is a clinical need for a vaccine to protect patients from chronic PA infection. This project involves using proteomic approaches to identify new vaccine antigens against P. aeruginosa and evaluating their host response. This ESR exploits our proteomic technology platform to identify a series of vaccine antigens by specifically identifying bacterial proteins involved in host cell binding; confirm expression during human infection by immunoproteomics; test efficacy as prophylactic vaccines in an acute infection model; and characterise mucosal response.
ESR2 – Novel T-cell based vaccines to fight ESKAPE infections globally
Organisation: Queens University Belfast (UK) Researcher: Paulina Zarodkiewicz Supervisor: Prof. Miguel Valvano Project description: We are employing a pipeline to identify proteins in the core proteome of multidrug antibiotic resistant ESKAPE pathogens, especially Klebsiella, Acinetobacter, and Enterobacter species. The ESR is developing a bioinformatics pipeline for in silico discovering of bacterial protein epitopes associated with most common human HTLA haplotypes from the core proteome, which can be used as T-cell antigens. The researcher then tests the long-term protectivity and memory response upon immunisation in mice and select the most appropriate candidates. Finally, the ESR will map the nature of the immune response against the most effective vaccine form (e.g. T- vs. B-cell responses).
ESR3 - Vaccine antigen identification against systemic infections
Organisation: Ludwig Maximilian University of Munich (Germany) Researcher: Océane Sadones Supervisor: Prof. Johannes Huebner Project description: ESKAPE bacteria are of great concern in the clinical setting due to their virulence and multiple antibiotic resistances. These pathogens are able to escape the biocidal effects of many antimicrobial agents and are major causes of nosocomial infections worldwide. Therefore, the development of non-antibiotic treatments such as vaccines is of great importance. In this context, the ESR focuses on methods to identify and characterise vaccine targets in enterococcus and staphylococcus, which are related to serious systemic infections. The ESR will identify carbohydrate and protein antigens targeted by opsonic antibodies; purify and characterise the antigens; conjugate protein and carbohydrate antigens for immunisation of rabbits; assess protective efficacy of rabbit sera and conjugates in established mouse models.
ESR4 - Structural and functional characterisation of novel vaccine antigens
Organisation/Institute: National Research Council, Institute of Biostructures and Bioimaging (Naples, Italy) Researcher: Eliza Kramarska Supervisor: Dr. Rita Berisio
Project description: Structure-based antigen design approach is a valid strategy for next-generation vaccine development. To this aim, the structural definition of the overall atomic-resolution structure of an antigen of its epitope regions is the driving force in the production of engineered antigens with improved immunological properties and provides biophysical tool that facilitate vaccine design and production. This project involves the use of a plethora of structural biology and biochemical approaches to characterise vaccine antigens against bacterial pathogens, including previously identified and patented efficacious Burkholderia antigens (UCD) and new Gram-positive and Gram-negative antigens, identified at UCD, QUB and LMU. The ESR will (i) clone, express & purify the antigens; (ii) characterise antigens by x-ray crystallography and spectroscopy; (iii) study protein-protein interactions by Surface Plasmon Resonance & Isothermal Titration Calorimetry; (iv) rationalise biological results using experimental and in silico analysis and identify mechanisms of action of identified antigens, and (v) apply data to design synthetic peptide epitopes. The ESR will involve secondments at CIC bioGUNE (Bilbao, Spain), EFPL (Lausanne, Switzerland) and IMXP (Gosselies, Belgium) for training in STD-NMR and in the development of synthetic peptide vaccines and analysis of T cell responses.
ESR5 - NMR MOLECULAR RECOGNITION OF ANTIGENS BY KEY RECEPTORS
Organisation/Institute: CIC bioGUNE | Center for Cooperative Research in Biosciences (Bilbao, Spain) Researcher: Unai Atxabal Supervisor: Prof. Jesús Jiménez-Barbero
Project description: The ESR is being trained in the study of molecular recognition events, with particular emphasis on the application of state-of-the-art NMR and molecular modelling techniques for the structural characterization of the complexes of different antigens with antibodies. In this project, the ESR (i) investigates conformations of the antigens, their analogues and their molecular mimics with emphasis on structural and functional information by NMR spectroscopic techniques; (ii) The experimental data will be combined with those obtained molecular modelling (docking and molecular dynamics) and other biophysical techniques (ITC).
ESR6 - Development of Toll-like Receptor-Directed adjuvants and incorporation in innovative vaccine formulations
Organisation/Institute: University of Milano Bicocca (Milan, Italy) Researcher: Ana Rita Franco Supervisor: Prof. Francesco Peri
Project description: Adjuvants are important components of vaccine formulation that optimise the protective immune response. Our group recently patented and published new small molecules that activate Toll-like Receptor 4 (TLR4), thus acting as immunostimulating agents and can be used as vaccine adjuvants. In this project the ESR: 1) projects and synthesizes new TLR4 agonists; 2) analyses the TLR4 activity on cells of natural compounds extracted from natural sources; 3) will formulate the adjuvant in the vaccine (in collaboration with Pfizer). Natural compounds will be also chemically modified to improve their pharmacodynamics and pharmacokinetic properties.
ESR7 - Elucidation of the host response to facilitate more effective vaccines
Organisation/Institute: CIC bioGUNE | Center for Cooperative Research in Biosciences (Bilbao, Spain) Researcher: Samuel Pasco Supervisor: Prof. Juan Anguita
Project description: Persistent pathogens have co-evolved with theirs hosts and developed strategies to modulate the immune responses of the host in order to avoid their clearance. These strategies include the ‘training’ of innate immune cells. The modulation of innate immune responses is particularly important in tissues in which the penetration of antibodies is poor, or the induction of antibodies responses (i.e. mucosal sites) is insufficient or inappropriate. The design of vaccine formulations that not only induce T and B cell responses but are also able to enhance innate immune responses (particularly phagocytosis) offers novel strategies to control infections, especially those that yield predominant antibody responses that are not particularly efficient. The ESR evaluates the comparative generation of immune responses against pathogens and symbionts as well as vaccine formulations against several microorganisms, including the mucosal pathogens enterohemorrhagic E. coli (EHEC) and Mycobacterium tuberculosis.
ESR8 - Production and characterisation of fusion protein antigens with a focus on increased solubility for large membrane proteins
Organisation/Institute: LIONEX GmBH (Braunschweig, Germany) Researcher: María García Bengoa Supervisor: Prof. Mahavir Singh
Project description: This project will focus on production and characterisation of fusion proteins with a high emphasis on increased solubility particularly for large membrane proteins and other biomarkers identified as vaccine candidates. The ESR will (i) be trained in the optimization of expression of high-quality proteins under ISO13458 certified conditions; (ii) clone vaccine candidates; (iii), fuse, express, purify in preparation for efficacy testing in different models; (iv) scale-up potential vaccine candidate expression using LIO GMP-compliant facilities and evaluate expression in several bacterial, yeast, baculovirus and eukaryotic systems; (v) characterise vaccine targets by the Octet platform analysis for studying receptor-ligand interactions.
ESR9 - Optimisation of host response and t-cell targeting
Organisation/Institute: Imperial College London (London, UK) Researcher: Franziska Pieper Supervisors: Prof. Rosemary Boyton and Prof. Danny Altmann
Project description: This ESR project is based in the busy molecular immunology lab of Prof. Rosemary Boyton and Prof. Daniel Altmann. The proposed studies will build on ongoing work in the lab to characterise the immunology of Pseudomonas aeruginosa (PA) infection – a cause of chronic lung infection of susceptible individuals such as those with bronchiectasis and cystic fibrosis. PA has genomic and transcriptomic adaptations that allow it to survive in the host lung, but the impact of these on immune system recognition have been only partially characterised. The first part of the project investigates PA adaptations on innate immune activation and antigen-presenting cell gene expression and function. During the next phase of the project, the ESR focuses on studying the impact of these changes on T cell antigen recognition, activation and polarisation. In the last stage of the project, this knowledge will be used to characterise protective and pathogenic immune mechanisms in lung infection models, including analysis in our ongoing vaccine programme.
ESR10 - Post-Exposure vaccination against multi-drug resistant tuberculosis
Organisation/Institute: St. George’s University of London (London, UK) Researcher: Emil Vergara Supervisor: Dr. Rajko Reljic
Project description: Tuberculosis is the single biggest killer among infectious diseases with 1.5 million deaths globally in 2018. Better measures are required at all levels to bring this dreadful disease under control, including better treatments, diagnostics and vaccines. The ominous rise in incidence of multidrug-resistant TB (MDR-TB) calls for further urgency in dealing with this global health threat. Post-exposure vaccination is an attractive approach to reducing disease incidence and transmission, particularly in the context of MDR-TB where the treatment options are limited. In this project, the ESR explores the potential of monoclonal antibodies (passive vaccination) and polymeric fusion protein antigens (active vaccination) to suppress an ongoing infection with MDR-TB. We have at our disposal newly built state-of-the-art CL3 containment facilities for working with Mycobacterium tuberculosis, including the highly advanced Biaera aerosol infection system for experimental TB. Our institute is one of the main UK centres for studying bacterial resistance for academic and clinical purposes.
ESR11 - Bicyclic peptide vaccine antigens
Organisation/Institute: École Polytechnique Fédérale de Lausanne (Lausanne, Switzerland) Researcher: Zsolt Bognár Supervisor: Prof. Christian Heinis
Project description: Bicyclic peptides can be exploited as drugs that bind to and modulate disease targets, or as mimics of antigens for vaccine development. Our laboratory is specialized on the in vitro evolution of bicyclic peptides by phage display. We have developed bicyclic peptides to a range of protein targets and are interested in translating them into therapeutics. ESR is focusing on (i) applying and further developing our bicyclic peptide phage display technology; (ii) identifying binders by NGS; (iii), chemically synthesizing bicyclic peptides by solid-phase peptide synthesis; (iv) characterising their binding properties and test their efficiency as prophylactic antigen; (v) developing and screening a library of bicyclic peptides; (vi), synthesising and characterising isolated bicyclic peptides, for testing of the best peptides as vaccines/ligands.
ESR12 - Optimisation of adjuvants for therapeutic vaccinations against respiratory infections
Organisation/Institute: University College Dublin (Dublin, Ireland) Researcher : Maite Sainz Supervisor: Assoc. Prof. Siobhán McClean
Project description: Effective vaccines require appropriate adjuvants to optimise the protective immune response. This project focuses on investigating novel adjuvants that elicit effective mucosal immune responses. The ES investigates host responses to chronic Pseudomonas aeruginosa infection, applies proteomic approaches to identify host receptors for novel vaccine antigens and assesses a series of novel BactiVax adjuvants and antigens to optimise a vaccine therapy for clearance of chronic infection. The ESR will also focus on developing methods to identify correlates of protection associated with clearance of chronic infection, on secondment in ImmunXperts (Gosselies, Belgium).
ESR13 - NANOCAPSULATED PEPTIDE-BASED Immunotherapeutics
Organisation/Institute: Eötvös Loránd University (Budapest, Hungary) Researcher: Chiara Bellini Supervisor: Dr. Kata Horváti
Project description: The most important issue in vaccine development is to balance safety with efficacy. The use of synthetic peptide-based vaccines which can trigger the desired immune response and can reflect on the antigenic variability is a safe approach, but the low immunogenicity needs to be addressed. In this project we investigate a new approach that combines vaccination, host-directed therapy and advanced vaccine/drug-delivery with the aim of improving treatment outcomes of antimicrobial resistant infection with a focus on Mycobacterium tuberculosis. The ESR focuses on the synthesis and characterisation of multicomponent peptide-based vaccine conjugates. To increase immunogenicity and overall bioavailability, conjugates will be formulated (polymeric nanoparticles, cyclodextrins, etc.) and the new candidates will be tested in vitro and in vivo in a murine model of tuberculosis.
ESR14 - Development of a Therapeutic vaccine against P. Aeruginosa
Organisation/Institute: Ludwig Maximilian University of Munich (Munich, Germany) Researcher: Enisa Smlatić Supervisor: Prof. Johannes Huebner
Project description: Pseudomonas aeruginosa is an important opportunistic pathogen causing a wide range of acute and chronic infections in immunocompromised patients. These pathogens pose increasing problems in the hospital setting due to the emergence of drug-resistant strains. This project will try to isolate and characterize novel monoclonal antibodies against P. aeruginosa. The ESR focuses on (i) isolating B-cells that produce opsonic and protective antibodies against bacterial pathogens; (ii) isolating and characterising novel monoclonal antibodies in vitro for affinity and opsonic activity; (iii) scaling-up promising monoclonal antibodies for efficacy testing in chronically infected mice in collaboration with UCD; (iv) identifying target antigens through secondments with partners CIC bioGUNE (Bilbao, Spain) and CNR-IBB (Naples, Italy).
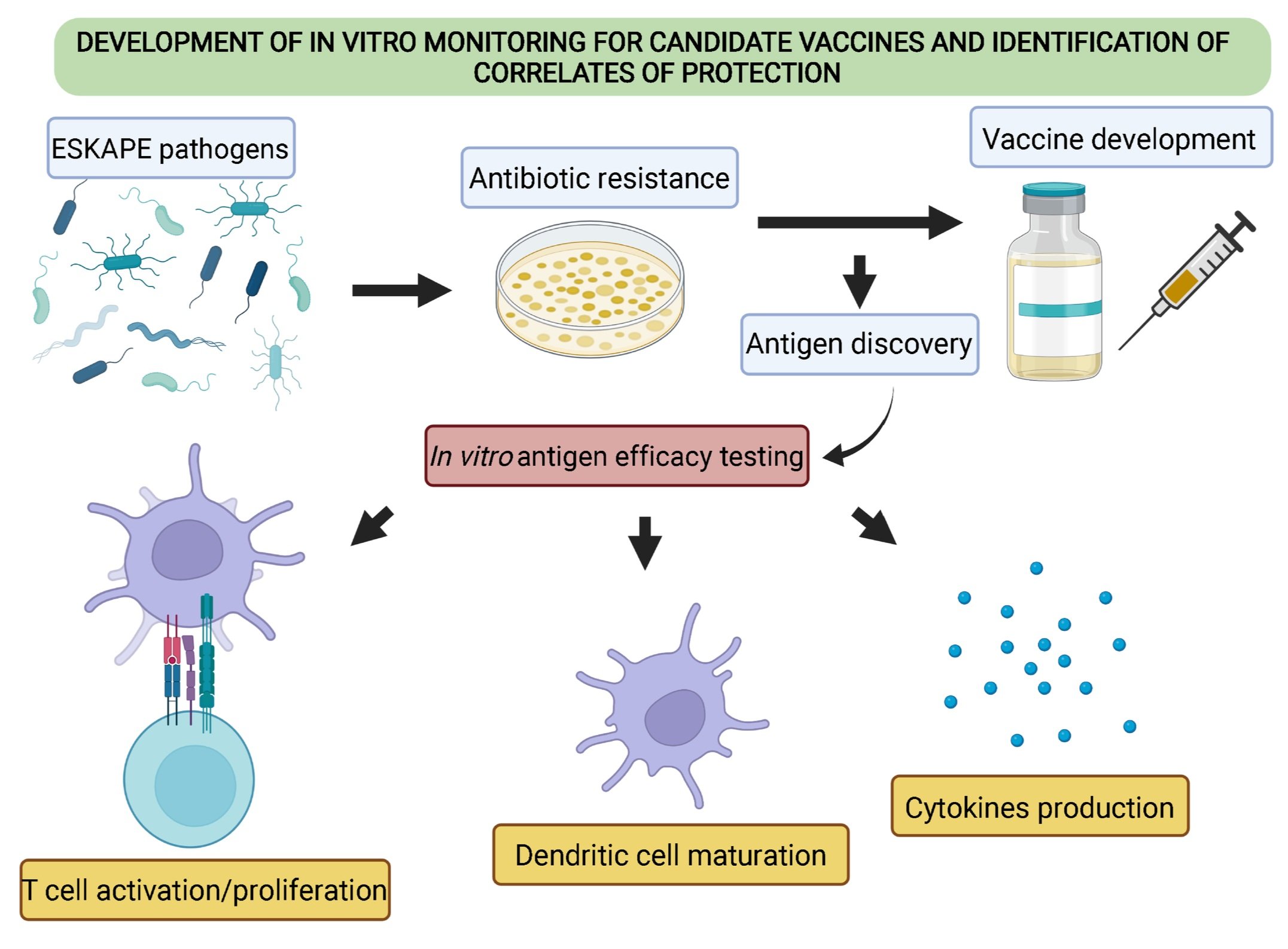
ESR15 - Development of in vitro monitoring for candidate vaccines and identification of correlates of protection
Organisation/Institute: ImmunXperts (Gosselies, Belgium) Researcher: Lorenzo Bossi Supervisor: Dr. Séverine Giltaire
Project description: This project will target selected ESKAPE pathogens (a group of multidrug resistant bacteria that are the leading cause of hospital infections globally, which “escape” biocidal action of antibiotics (3), e.g. Pseudomonas aeruginosa, staphylococci, enterococci). The ESR will be trained at IMXP in immunoanalytical platforms such as cytokine analysis (ELISA, HTRF and Luminex), T and B cell ELISPOT and multicolour flow cytometry for the evaluation of different phenotypic populations in combination with the secretion of cytokines. The ESR is focusing on optimising a suite of in vitro assays to evaluate candidate antigens/adjuvant combinations (identified in WP1) using human peripheral blood cells from healthy donors (male and female) and to identify correlates of protection in patient samples. ESR15, on secondment at St Georges Hospital, University of London (SGUL), will perform in vivo studies to evaluate Th17 response giving maximal protection against mucosal pathogens and will be trained in identification of host receptors using a proteomic approach at University College Dublin (UCD).